Obscurin proteins are a diverse group of muscle specific and ubiquitously expressed proteins. We and others have demonstrated that obscurin proteins play important roles for the architecture of various cellular membrane systems, like the sarcoplasmic reticulum (SR) and the muscular sarcolemma. In addition, obscurin proteins have been shown to influence the protein turnover of their interaction partners, and to play a role in muscle specific signaling pathways and - indirectly - for cellular calcium handling.
The obscurin protein family consists of four members:
- the giant muscle specific obscurin proteins (Obscurin-A & Obscurin-B isoforms)
- the obscurin kinase protein (Obkin)
- the striated muscle preferentially expressed gene/protein - SPEG/APEG
- and the ubiquitously expressed obscurin-like 1 protein (Obsl1)
Obscurin
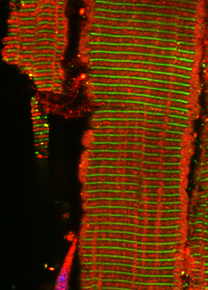
The giant obscurin protein is thought to link the contractile apparatus in muscle cells with the sarcoplasmic reticulum (SR), a membrane system specialized in the storage of calcium. Lack of obscurin in cross-striated muscle cells leads to changes in longitudinal SR architecture. Changes in SR architecture in obscurin knockout mice were also associated with alterations in several SR or SR-associated proteins, such as ankyrin-2 and beta-spectrin. As a consequence, obscurin knockout mice display centralized nuclei in skeletal muscles as a sign of mild myopathy, but have normal sarcomeric structure and preserved muscle function.
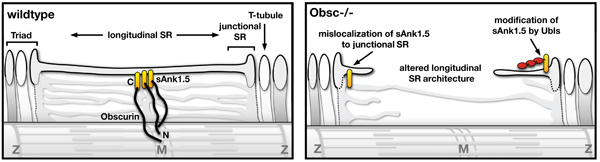 Model of obscurin functions for the SR architecture in muscles.
Model of obscurin functions for the SR architecture in muscles.
Published in the Journal of Cell Science. Follow the link for access to the full manuscript.
|
In a contribution to a collaborative research project headed by Dr. Sorrentino's group at the University of Padua, Italy, we investigated how obscurin is also important for the structure and integrity of the specialized skeletal muscle membrane - the sarcolemma.
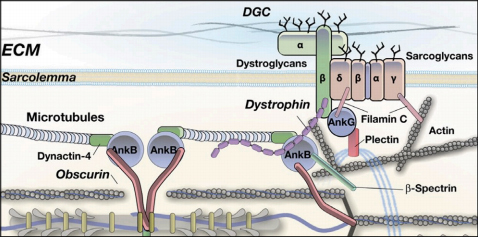 Model of obscurin functions for the skeletal muscle sarcolemma membrane.
Model of obscurin functions for the skeletal muscle sarcolemma membrane.
Published in the Journal of Cell Biology. Follow the link for access to the full manuscript.
|
We also elucidated the mechanism on how obscurin influences the protein turnover of one of its binding partners - small ankyrin 1.5 (sAnk1.5) in heart and skeletal muscle cells. Intriguingly, degradation of sAnk1.5 is further regulated by posttranslational modifications by nedd8, ubiquitin and by acetylation of C-terminal lysine residues.
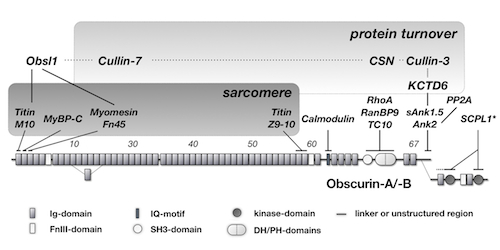
Obscurin domain layout and summary of known binding partners.
Published in Molecular Biology of the Cell. Follow the link for access to the full manuscript.
|
Redundancy among obscurin protein family members
Obscurin knockouts only present with a mild form of myopathy. We hypothesized that the close homologue obscurin-like 1 (Obsl1) compensates for the loss of obscurin in skeletal muscles. To investigate functional redundancy among obscurin protein family members, we generated and characterized Obsl1 knockout mice.
Global loss of Obsl1 is embryonically lethal, necessitating the generation of muscle specific Obsl1 knockouts. Similar to obscurin knockouts, muscle-specific Obsl1 knockouts present with a benign muscle phenotype. Only loss of both proteins, obscurin and Obsl1 revealed their role for muscle sarcolemma, SR and muscle metabolism.
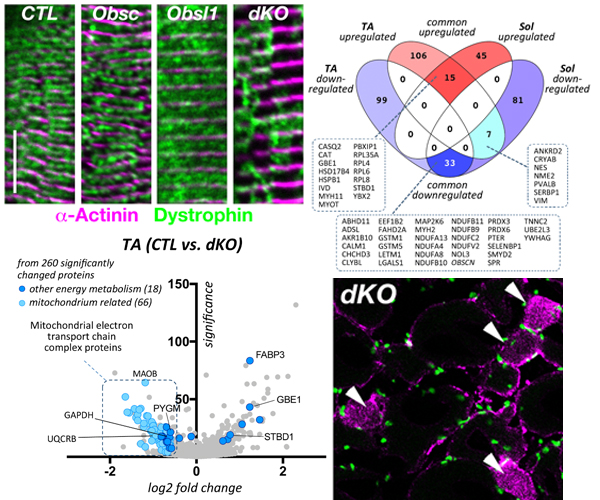
Loss of both proteins, obscurin and Obsl1 results in increased sarcolemmal instability, altered SR structure/function and metabolic changes.
This work was published in the journal Communications Biology. Follow the link for access to the full manuscript.
|
Unlocking secrets of obscurin and SPEG kinase domains
Three members of the obscurin protein family contain tandem kinase domains with important signaling functions for cardiac and striated muscles. These members are the B-splice isoform of the giant protein obscurin, the small obscurin-associated kinase splice isoform, and the closely related striated muscle enriched protein kinase (SPEG). While mutations in and around the kinase domains have been linked to a number of cardiomyopathies in humans, functions of the kinases and their regulation remain enigmatic.
We investigated a role for low complexity inter-kinase region C-terminal to kinase domain 1 in obscurin and SPEG. We uncovered a novel autophosphorylation of obscurin in a part of the interkainse region that bears sequence resemblance to a known substrate of SPEG kinase. Our manuscript also highlights structural similarities of obscurin kinase 1 to Death-associated kinases, allowing for homology modeling.
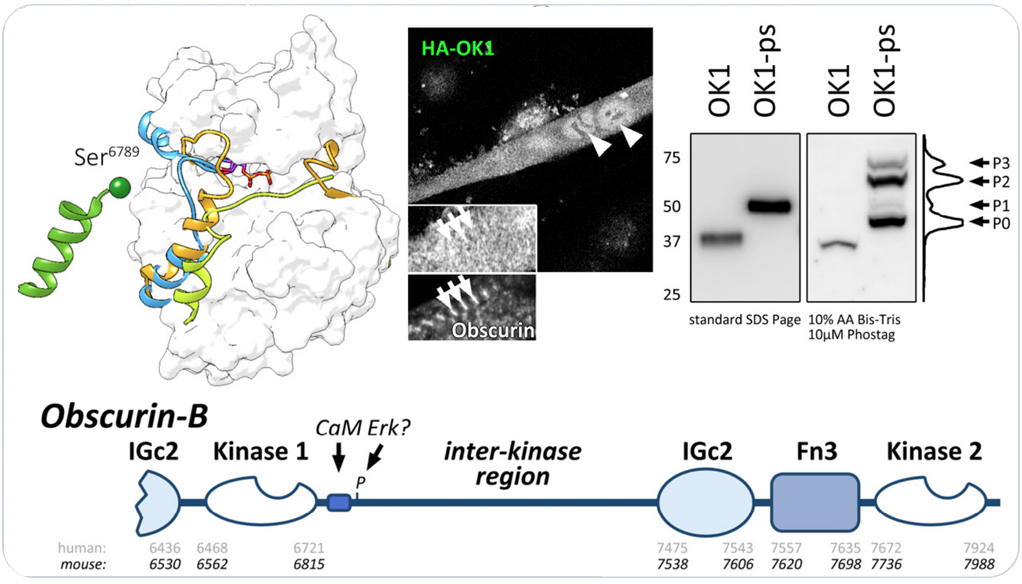
Obscurin kinase domain 1 autophosphorylates the protein in its C-terminal the interkinase region. Follow the link for access to the full manuscript that investigates obscurin and SPEG kinase biology.
|
Cardiac roles of Obscurin and Obsl1
In our recent preprint, we explored cardiac roles for obscurin and Obsl1. Loss of either gene/protein was tolerated in mice. However, combined loss of obscurin and Obsl1 resulted in premature death of mice, characterized by normal systolic function at baseline, but impaired diastolic function. Stressed hearts also display loss of cardiac reserve and systolic impairment. Double knockout (dKO) mice displayed diastolic heart failure, reminiscent of patients suffering from heart failure with preserved ejection fraction (HFpEF).
On the molecular level, dKO hearts display altered sarcoplasmic reticulum architecture and function, impacting calcium cycling. In addition, hearts show altered cardiac metabolism and impaired mitophagy as a result of loss of Obsl1 binding to Atg4d. Mitochondria structure is also affected, as levels of Micos protein Chchd3 are down regulated in dKO hearts.
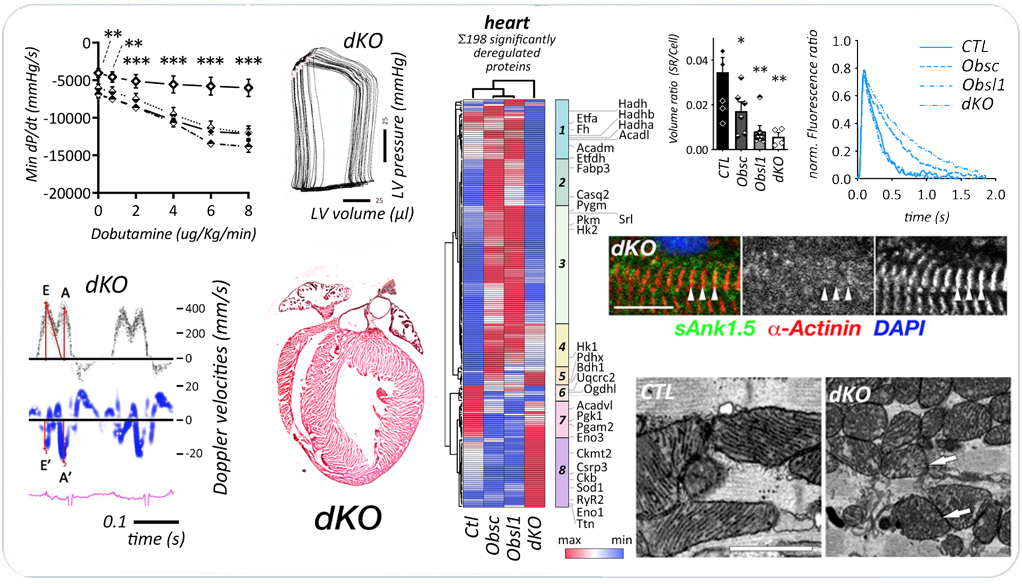
Combined loss of obscurin and Obsl1 in hearts results in diastolic heart failure. The preprint is available on bioRxiv doi:10.1101/2022.08.24.505098..
|
Future Directions
We continue to investigate cardiac roles for obscurin and Obsl1, and molecular mechnanisms for the development of diastolic heart failure and heart failure with preserved ejection fraction (HFpEF).
Related manuscripts
- PREPRINT: Combined loss of obscurin and obscurin-like 1 in murine hearts results in diastolic dysfunction, altered metabolism and deregulated mitophagy. Fujita K, Desmond P, Marrocco V, Blondelle J, Sotak M, Rajan M, Esteve E, Chan Y, Clark M, Gu Y, Dalton N, Ghassemian M, Do A, Klos M, Borgeson E, Peterson K, Shekh F, Lange S. bioRxiv. 2022 doi:10.1101/2022.08.24.505098.
- Exploring Obscurin and SPEG Kinase Biology. Fleming JR, Rani A, Kraft J, Zenker S, Börgeson E and Lange S. JCM. 2021 doi:10.3390/jcm10050984.
- Murine obscurin and Obsl1 have functionally redundant roles in sarcolemmal integrity, sarcoplasmic reticulum organization, and muscle metabolism.
Blondelle J, Marrocco V, Clark M, Desmond P, Myers S, Nguyen J, Wright M, Bremner S, Pierantozzi E, Ward S, Estève E, Sorrentino V, Ghassemian M, Lange S. Comms Bio 2019
- Obscurin is required for ankyrinB-dependent dystrophin localization and sarcolemma integrity.
Randazzo D, Giacomello E, Lorenzini S, Rossi D, Pierantozzi E, Blaauw B, Reggiani C, Lange S, Peter AK, Chen J, Sorrentino V. PMID: 23420875
- Obscurin and KCTD6 regulate cullin-dependent small ankyrin-1 (sAnk1.5) protein turnover.
Lange S, Perera S, Teh P, Chen J. PMID: 22573887
- Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum.
Lange S, Ouyang K, Meyer G, Cui L, Cheng H, Lieber RL, Chen J. PMID: 19584095
- Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: implications for hereditary myopathies.
Fukuzawa A, Lange S, Holt M, Vihola A, Carmignac V, Ferreiro A, Udd B, Gautel M. PMID: 18477606
- From A to Z and back? Multicompartment proteins in the sarcomere.
Lange S, Ehler E, Gautel M. PMID: 16337382
Collaborators on this project
- Dr. Mathias Gautel, King's College London, UK
- Dr. Vincenzo Sorrentino at the University of Siena, Italy
- Dr. Isabelle Richard at Genethon in Paris
- Dr. Bjarne Udd, from the University of Helsinki in Finland
- Dr. Guy Benian of Emory University
|